Tutorial: using NeurEco GUI on a Tabular Classification problem
Tutorial: using NeurEco GUI on a Tabular Classification problem#
This section uses the test case Gene expression cancer RNA sequence. This test case can be selected directly from the template window of the GUI.
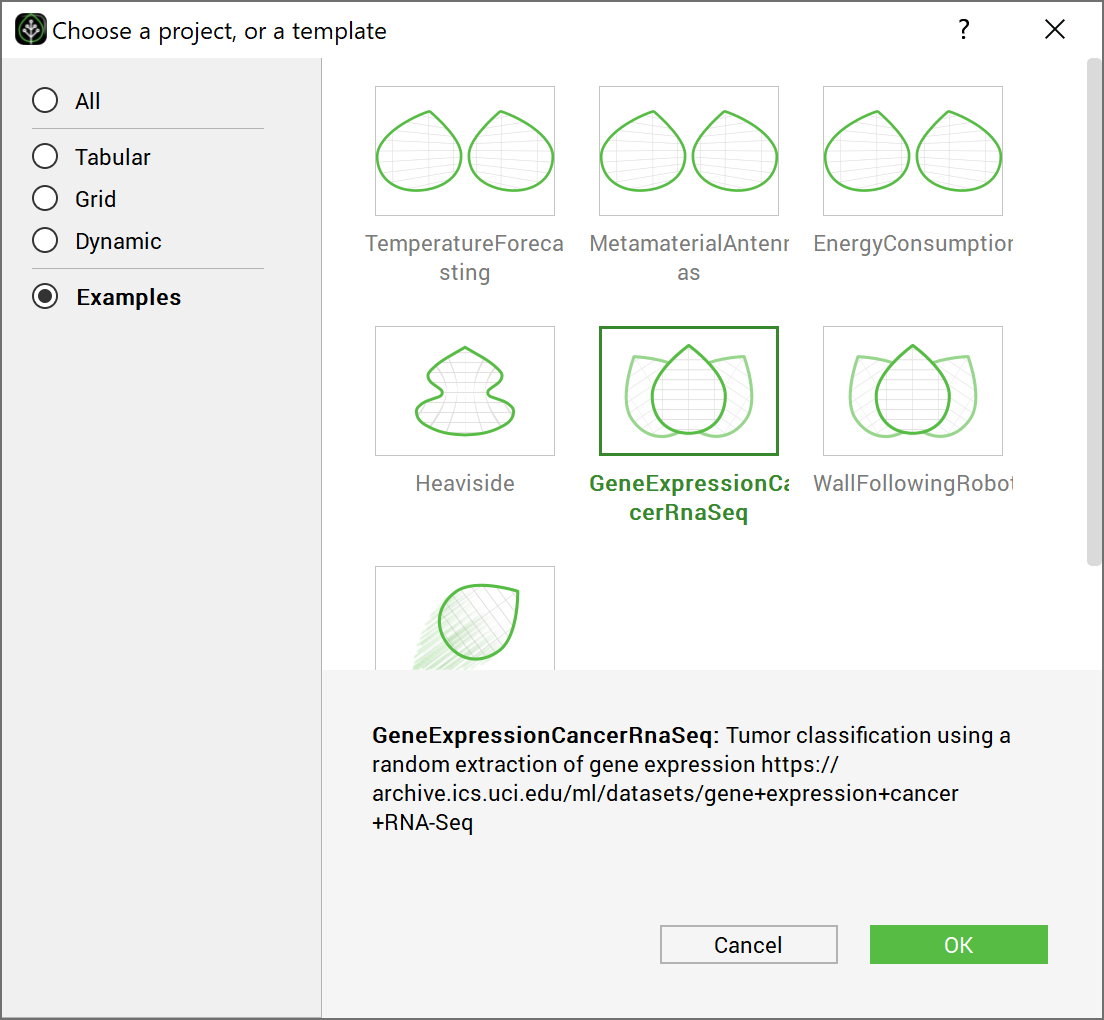
Choosing the test case GeneCancer directly from the GUI examples#
Create an empty directory (GeneCancer Example), extract the Gene expression cancer RNA sequence test case data there. The GUI automatically extracts the data and creates the project in the chosen directory. The created directory will contain the following files:
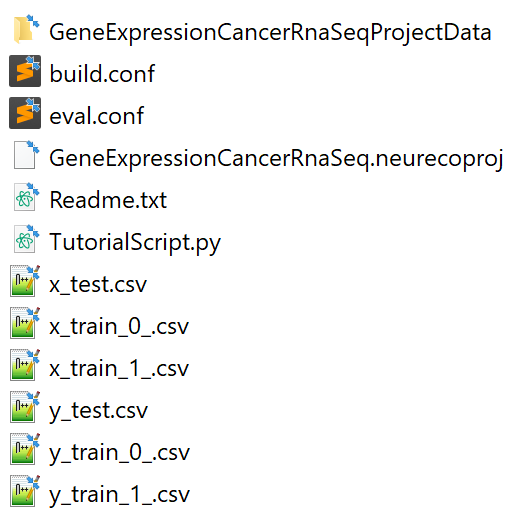
Content of the test case GeneCancer from the GUI#
The GeneCancer directory is used by the GUI alongside the CSV data files. The rest is used by the other NeurEco interfaces.
Note
To create the GUI project without using the template window, create a new directory called GeneCancer and copy the data CSV files into it. Go to the File menu, and click New, then choose the Tabular solution and the Classification template. Choose the name of the project and the name of the model as: GeneCancerTutorial and GeneCancer and click ok.
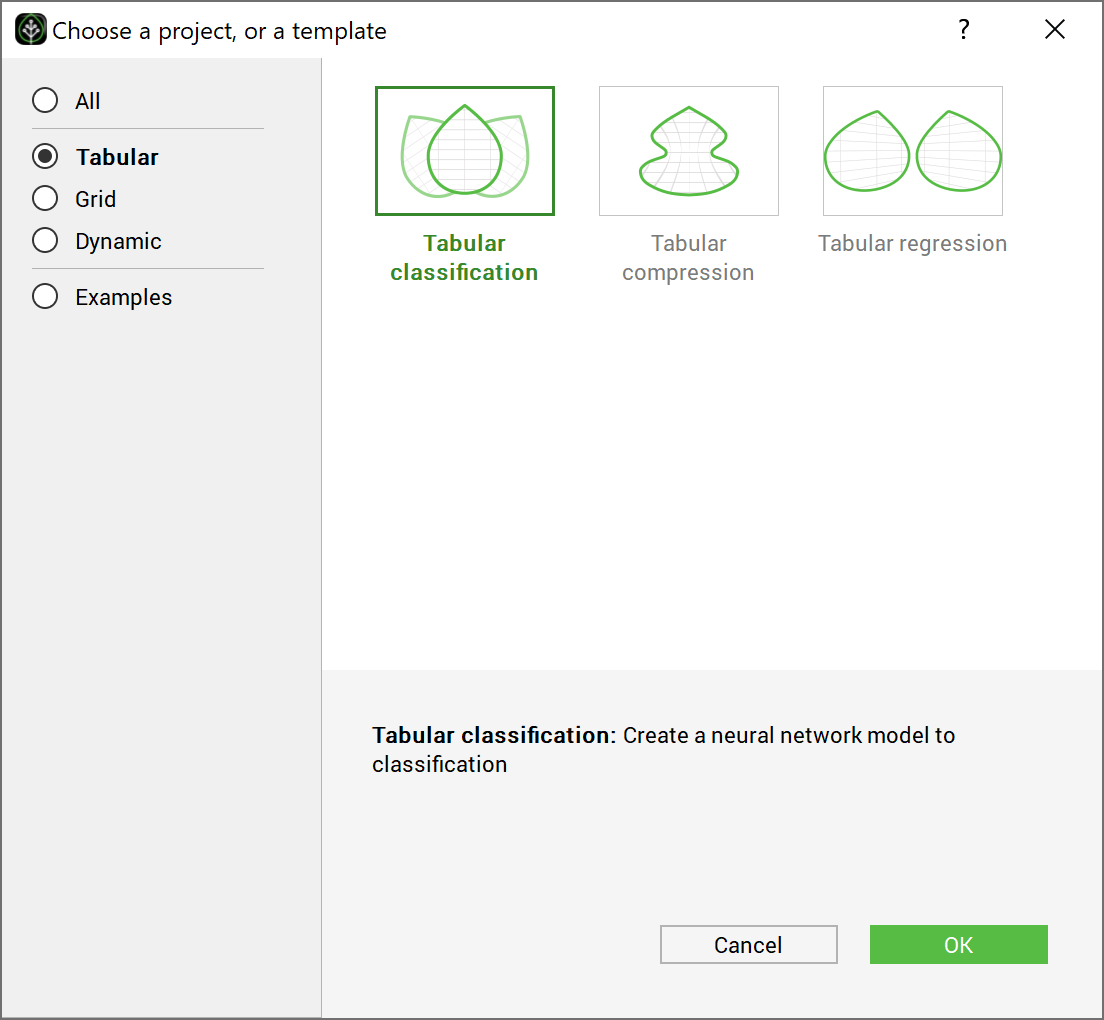
Choosing the test case GeneCancer directly from the GUI examples 2#
The main window looks as follows at this stage:
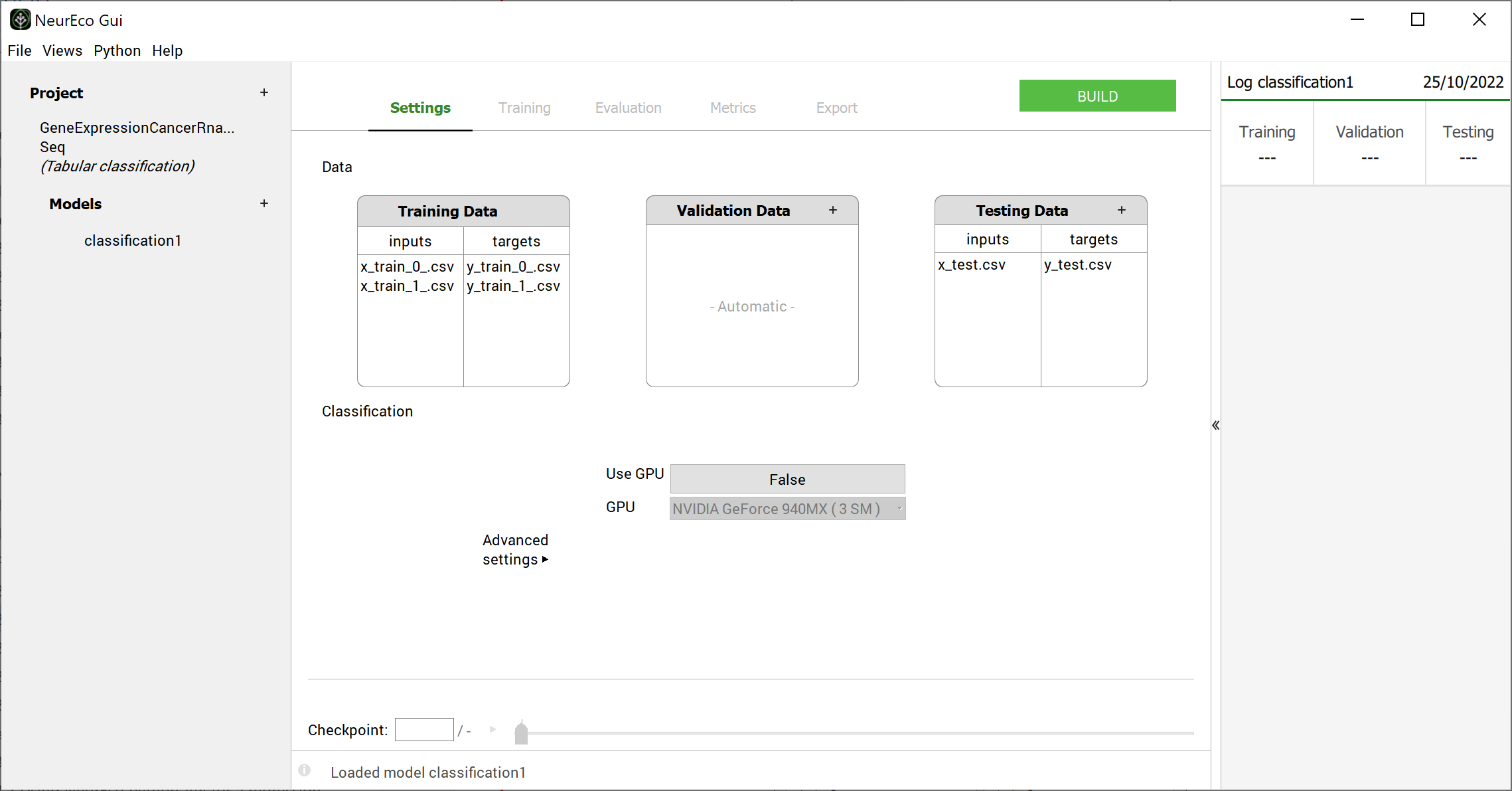
Main window initial look after extracting the data: test case - GeneCancer#
Note
For the Classification cases, NeurEco expects the outputs to be one hot encoded. It does not accept labeled outputs. See Data preparation for NeurEco Classification with GUI.
To build a model:
Adjust the Settings (add some data for the learning, validation or test, change one or more building parameters (see Build parameters). Here, for Gene expression cancer RNA sequence test case, the Settings keep their default values.
Click on the Build button
During the build NeurEco saves the intermediate modes to the checkpoint file. In term of performance, every new model in the checkpoint is an improvement of the previous one. Note that at the end of the build, the last model in the checkpoint corresponds to the final mode.
Any intermediate model can be used as if it was the final model: it can be evaluated on the new sets of data, exported, etc. Use the checkpoint slider to select a specific intermediate model. When an intermediate model is selected, the GUI updates the plot of the network architecture, the plot of reference vs prediction and the Sensitivity analysis plot (see Sensitivity analysis for Tabular solutions).
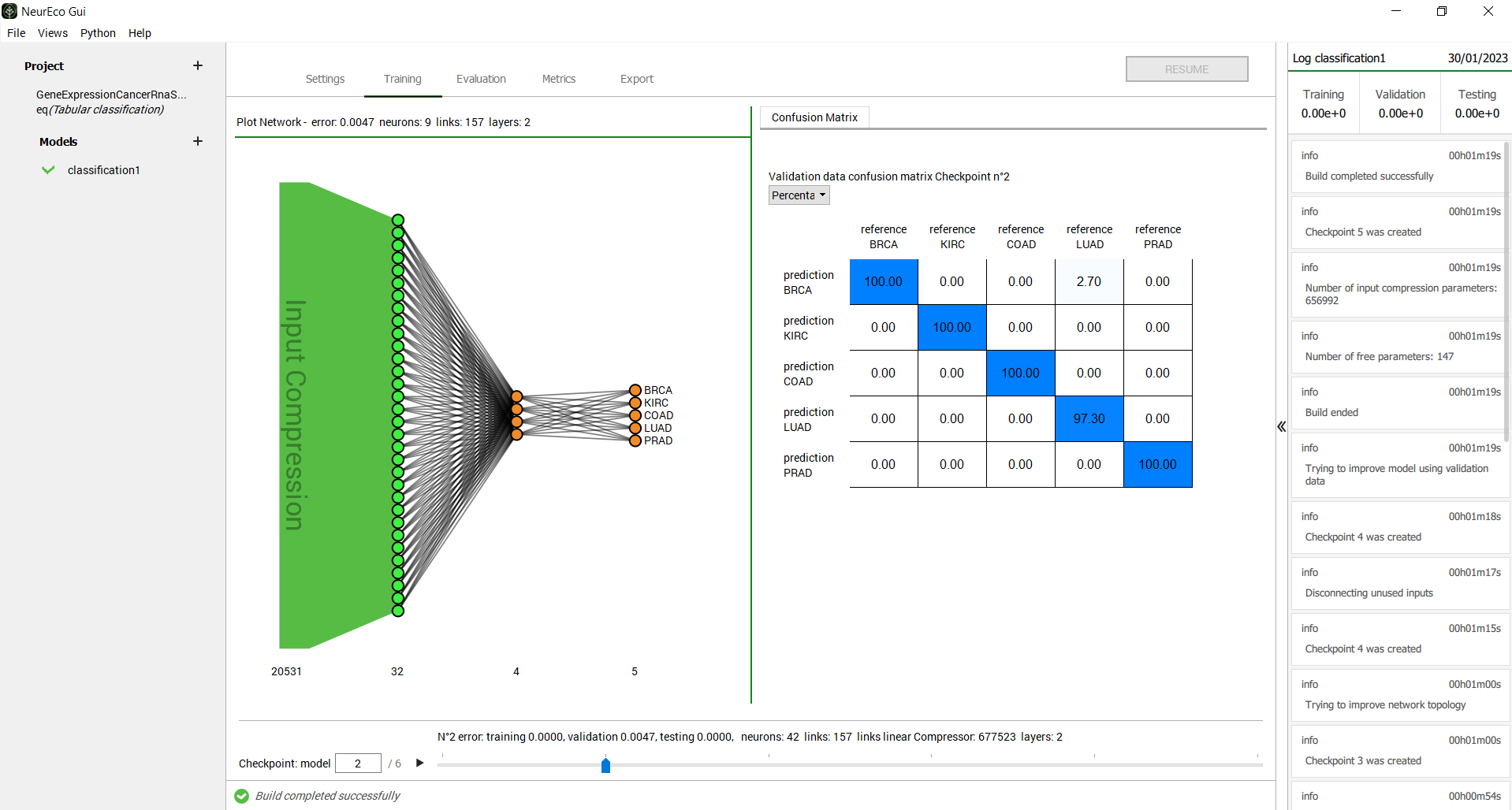
GUI operations: selecting an intermediate model: test case - GeneCancer#
Note
The number of links shown by the GUI is the number of trainable parameters in the network. Each link between two neurons represents a parameter, plus there are the bias parameters not shown on the network plots.
The Evaluation panel allows a user to load extra sets of data to evaluate the model on and then to export the results in a csv or npy format (see Evaluate NeurEco Classification model with GUI).
The Metrics panel allows a user to calculate a set of metrics (see Metrics for the Tabular Classification model with GUI). For the Classification problems these metrics looks as shown in the figure below:
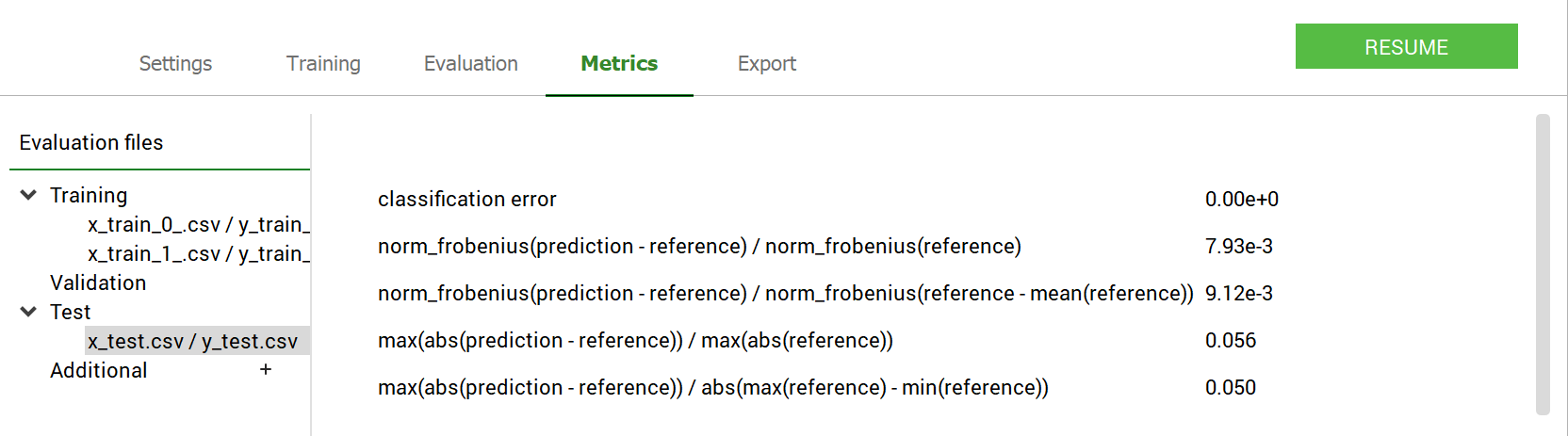
GUI operations: Extracting the metrics: test case - GeneCancer#
To export the model (see Export NeurEco Classification model with GUI, embed license is required for export to formats different from NeurEco model):
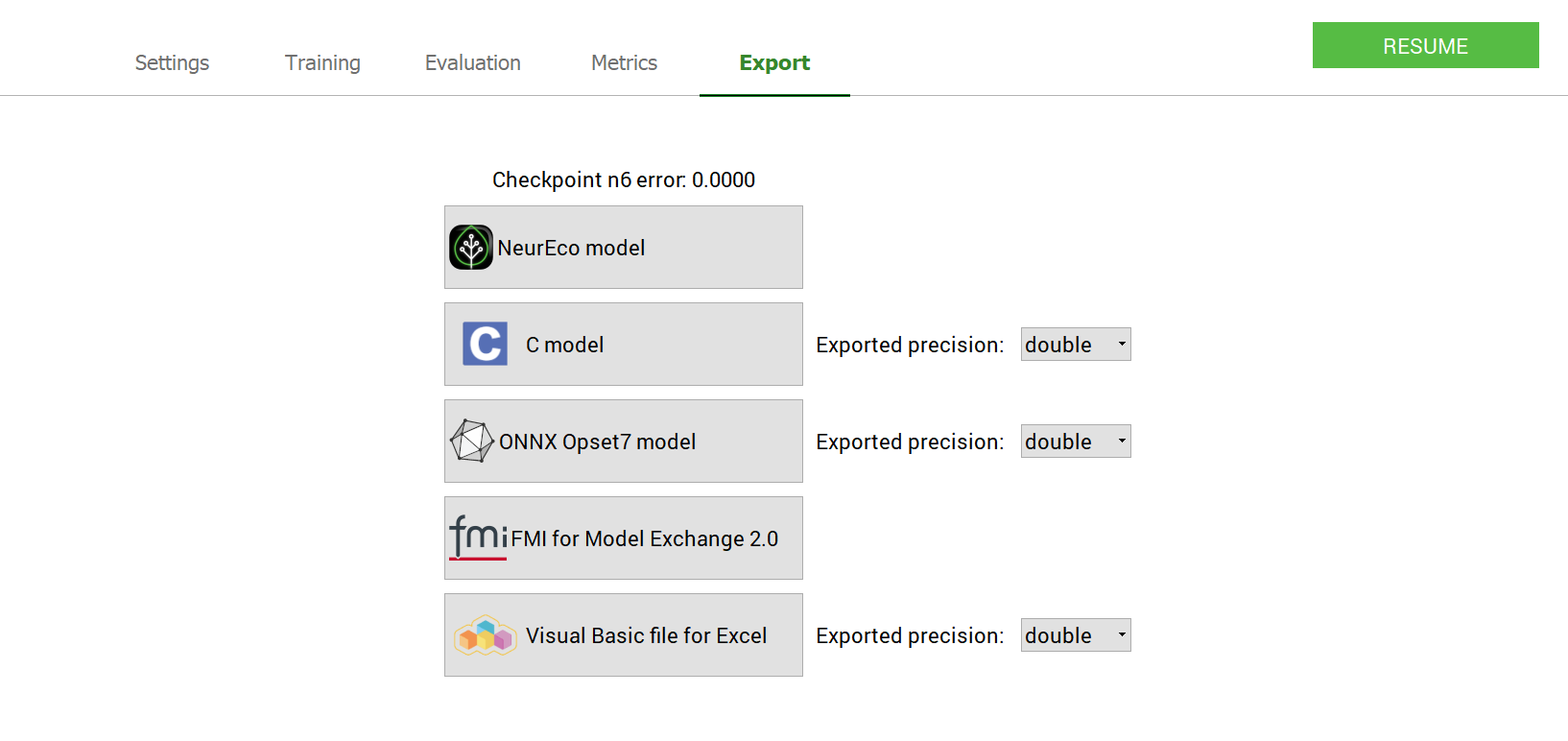
GUI operations: Exporting a model : test case - GeneCancer#
To create a Python script reproducing the main parts of the GUI project (see Export Tabular Classification from the GUI to the Python API):
Go to Python/Export NeurEco to Python in the menu bar of the GUI
Choose which parts of the project to export to a Python script
Select the destination where to save the script
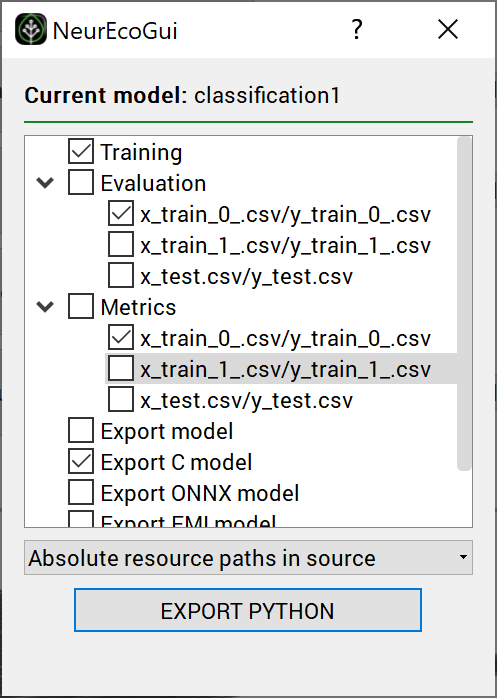
GUI operations: Exporting a python script : test case - GeneCancer#
Warning
To be able to use the script exported from the GUI, the NeurEco Python API package has to be installed on your computer.
